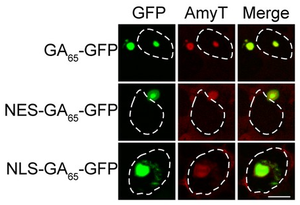
Repetitive genomic regions are known to expand across generations, due to errors during their replication, and cause a series of - mainly - neurological diseases collectively called repeat expansion diseases. One of these repetitive genomic regions is the GGGGCC (G4C2) locus in the C9orf72 gene. Its expansion is linked with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). It is still unclear, however, whether the cytotoxic effects of the repeat-expansion are emerging from the G4C2 mRNA, or from the results of its translation. Frederic Frottin from F. Ulrich Hartl’s group at the Max Planck Institute of Biochemistry and Mark S Hipp at the University Medical Center Groningen tried to dissect the issue in their paper published in the eLife journal. In order to distinguish the effects of the mRNA from the ones of the resulting protein, the researchers designed a construct that produces a poly-GA containing protein from a non-G4C2 RNA.They then used a Nuclear Export Signal (NES) and Nuclear Localization Signal (NLS) to localise the construct on either side of the nuclear envelope. Both constructs formed inclusions in their respective compartments which were stained by Amytracker 680, confirming the presence of amyloid structures within them. Leveraging this system, the authors were able to shed light on the toxic effects of the G4C2 expansion in the C9orf72 gene. Their work reveals that the expression of cytoplasmic poly-GA has only modest effects on cell viability, and that nuclear poly-GA proteins, however, are more toxic and impair nucleolar protein quality control and protein biosynthesis.
Image: Cytoplasmic and nuclear aggregates in HEK293 cells stained with Amytracker 680 (AmyT, red). Poly-GA DPRs were visualised by GFP fluorescence (green). White dashed lines delineate the nucleus based on DAPI staining. Image from Figure 1D by Frottin, F. et al. (2021) eLife, 10, e62718 (CC BY 4.0).
