Tauopathies, including Alzheimer's disease (AD) and Pick’s disease, are characterized by the aggregation of tau proteins into insoluble filaments. Traditional cell-based assays modelling tau seeding often rely on mutant or truncated tau, limiting their relevance to sporadic tauopathies. In a study published in the Journal of Biological Chemistry, Huang and McEwan developed a biosensor assay based on HEK293 cell lines stably over-expressing full-length, wild-type tau isoforms to more accurately model disease conditions.
They demonstrated that wild-type tau, without disease-associated mutations, is sufficient to develop sensitive and disease-relevant seeding assays when using brain-derived tau aggregates. Moreover, they showed that the aggregates in these cells can be propagated across passages, and the system can be used to generate clonal cell lines that show sustained AD-like, hyperphosphorylated tau aggregates.
By comparing HEK293 cells expressing wild-type and tau carrying the disease associated P301S mutation, the team showed how only in the former the aggregation is isoform-specific. Additionally, while recombinant wild-type Tau forms seed-competent aggregates, P301S Tau aggregated in vitro is unable to seed wild-type Tau aggregation.
Throughout their study, the researchers used Amytracker 680 alongside phosphorylation-specific antibodies to confirm the aggregation state of their constructs, emphasizing Amytracker’s utility in visualizing pathological tau conformations.
This work underscores the potential of using wild-type tau-expressing cell lines combined with sensitive detection methods like Amytracker to study the propagation of disease-relevant tau assemblies, offering a valuable platform for future therapeutic screening and mechanistic studies in tauopathies.
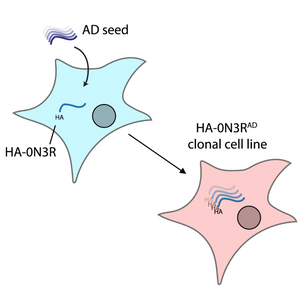
Image: A diagram depicting how AD-brain-derived tau seeds can propagate in HEK293 cells expressing HA-Tau (HA-0N3R) and be retained across generations (HA-0N3RAD cell line). Image adapted from figure 3A of Huang M. and McEwan W.A. (2025) J Biol Chem 301(3):108245 (CC BY 4.0).
