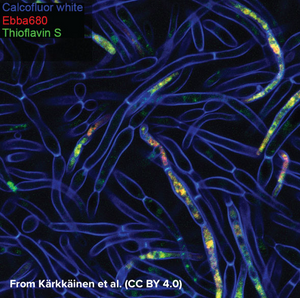
Candida albicans is a commensal fungus that lives among the gut flora of 40 to 60% of healthy individuals, where it presents as ovoid “yeast” cells. Changes in its environment can trigger a switch towards the formation of multicellular hyphae which is thought to be a crucial step for the virulence of the fungus as intertwined hyphal filaments enable the formation of a biofilm. This means that, through biofilm formation, C. albicans can become an opportunistic pathogen and cause infections, which is particularly problematic in immunocompromised patients. When forming a biofilm, the fungus surrounds itself with “fungal superglue” - an extracellular matrix (ECM) of β-1,3-glucans, lipids, and nucleic acids - that protects it not only against the immune system but also from antifungal drugs. This makes treatment of C. albicans infections particularly challenging with mortality rates of up to 40% in cases of systemic candidiasis. The limited number of detection methods for C. albicans further emphasises the need for technical development in this area. While biofilms can be measured by biomass or metabolic activity, distinguishing between growth states is hard, although critical for identifying active disease. Kärkkäinen et al. from AIMES at Karolinska Institutet in Sweden took on the challenge to try to identify specific markers for the biofilm status using fluorescent probes. The researchers used a range of fluorescent probes for detailed analysis of yeast cells and hyphae. Fluorescently tagged Concavalin A which is a Carbohydrate-Binding-Protein and Calcofluor white which binds to cellulose and chitin were used to label the cell wall. DAPI was used as a nuclear stain, MDY-64 was used to label cell membrane and vacuoles and Thioflavin S was used to label densely packed and dispersed amyloids. Whereas the use of these labels gave a very detailed view on the anatomy of cellular yeast and yeast hyphae in biofilms, no clear cues for differentiation of yeast cells and biofilm was found that didn’t involve microscopic investigation. As common markers failed to differentiate yeast cells from biofilms, the researchers tested if EbbaBiolight 680 can be used to discern between cell wall polysaccharides and thus differentiate between cellular yeast and biofilm. Initially, the researchers tested if EbbaBiolight 680 binds to purified components of the yeast cell wall and found that EbbaBiolight 680 is unable to detect mannans, but binds to β-glucans and chitin. Moreover, the researchers found that the emission spectrum of EbbaBiolight 680 bound to β-glucans differs from the emission spectrum of EbbaBiolight 680 bound to chitin, suggesting that EbbaBiolight 680 might be used to distinguish between the two. Therefore, the researchers used EbbaBiolight 680 to label live-cultures of C. albicans as a next step. Interestingly, they found that the signal from the cell walls was relatively weak compared to the bright signal obtained from the intracellular amyloids (Image from: Kärkkäinen, E. et al. (2022) Frontiers in Cellular- and Infection Microbiology 12, 981454. CC BY 4.0). Furthermore, significant differences were found in the emission spectra of EbbaBiolight 680 bound to intracellular amyloid structures in yeast cells compared to cells growing in a biofilm . EbbaBiolight 680 bound to amyloids in yeast cells showed a broad emission spectrum with two peaks: one at 498 nm and one at 614 nm whereas EbbaBiolight 680 bound to amyloids in yeast biofilm showed an emission spectrum with one peak at 634 nm. Thus, the ratio of the fluorescence intensity (RFU) at 498 nm and at 634 nm was used as a measure to tell the difference between cellular yeast and yeast biofilm. In summary, the publication by Kärkkäinen et al. shows the potential for EbbaBiolight to differentiate between yeast cells and yeast biofilm and thereby solve a clinical problem providing means of differentiating between the commensal and pathogenic lifestyle of a microorganism.
